Researchers at the Indian Institute of Science (IISc) have developed a method to rapidly identify and check whether a disease-causing bacterium is alive or dead.
It uses Raman spectroscopy, a technique that is usually used to identify chemical bonds in materials, to recognize bacteria and check for their viability, or their state of being alive.
"The uniqueness of this study is the rapidity and sensitivity of the method, and its potential for being modified into a bed-side, table-top device for diagnosis," says Srividya Kumar, a former Ph.D. student at the Department of Inorganic and Physical Chemistry, and first author of the study published in the journal Analytical and Bioanalytical Chemistry.
Quick detection of the disease-causing agent or pathogen, as well as checking for whether it is alive or not in patient samples are key to treating infectious diseases. Identifying the pathogen's viability also helps physicians decide the dose of antibiotics to be prescribed, and reduces the possibility of overprescription that can lead to antibiotic resistance.
Infectious bacteria are usually identified by techniques such as culturing—growing them on a nutrient medium in a Petri dish. It can take about two to three days to track their growth and confirm whether they are dead or alive. In addition, bacteria that are difficult to grow in lab cultures often go undetected by conventional methods. More sophisticated methods such as Polymerase Chain Reaction (PCR) create a genetic profile of the microbe, but cannot tell whether it is alive or dead.
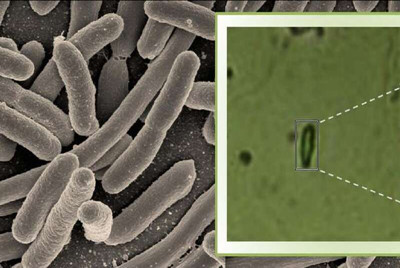
Raman spectroscopy is used widely in the field of chemistry to probe the structure of molecules. In this study, however, Kumar and her colleagues have combined this approach with advanced microscopy to check for the presence of bacteria. They used this technique to detect whether the tuberculosis bacterium (Mycobacterium tuberculosis) was present in a sample medium that resembles sputum. They also tested samples containing five other microbes.
Using the microspectroscope, the researchers bounced a laser beam off the sample and captured the light scattered by the sample in the form of a spectrum, which varies with the biochemical composition of the bacterium. Each bacterial species generates a unique spectrum with specific values for intensity and wavelength position of the scattered light, depending on the type of chemical bonds present inside the bacterial cell. The technique was sensitive enough to generate a spectrum even for a single bacterial cell.
The material that is used for mounting the bacteria in this technique, known as the substrate, is also crucial because some substrates can add noise to the biochemical profile. The researchers developed a neutral, aluminium-based thin film substrate which does not generate background signals that may interfere with the spectra.
Using this method, the researchers were not only able to identify the bacterium but were also able to differentiate between living and dead bacterial cells—since the chemical composition of the two differs—within two to three hours after sample collection.
As each bacterial species gives rise to a unique spectrum, it might be possible to eventually build a database of pathogens that can be used in clinical diagnosis.
"Though we have validated the method for tuberculosis, the methodology can be extended to any type of bacterial infection," says Deepak Saini, associate professor in the Department of Molecular Reproduction, Development and Genetics, IISc, and one of the senior authors of the study.

