Scientists at the University of Maryland (UMD) and the National Institute of Standards and Technology (NIST) have discovered a way to detect the molecular mechanism by which 5HT3A, a serotonin receptor located at the neuron synapse, is activated. Having a molecular model of this activation will allow the testing of pharmaceuticals inhibitors using computer models instead of traditional experiments, potentially reducing the cost and time of screening new drugs.
The study led by Jeffery Klauda, a professor of Chemical and Biomolecular Engineering (ChBE) at the University of Maryland (UMD), was published in the Proceedings of the National Academy of Sciences (PNAS) today. Nicholas Guros, a ChBE graduate research assistant, served as first author on the study and was co-advised by Arvind Balijepalli, a mechanical engineer at NIST.
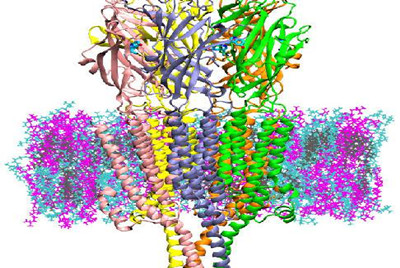
5HT3A is a type of protein known as an ion channel, implicated in depression, anxiety and targeted to reduce chemotherapy-induced nausea. It regulates the flow of ions at the post-synaptic cleft of neurons. The authors performed computer simulations of 5HT3Afor up to 20 microseconds—much longer than past simulations performed for
"If you think of molecular dynamics (MD) as a molecular microscope, this means we're able to observe the mechanism of activation for x10 longer than past," said Guros. "New computational capabilities can provide the power to observe the molecular activation of complex proteins at significantly greater timescales. The longer this timescale, the better validation we can have of experimental results to demonstrate MD models are a robust method to study protein and test pharmaceuticals."
The activation and desensitization of proteins like 5HT3A occur over milliseconds, so while closer to the true timescale, scientists are still somewhat behind observing the entire transport cycle of the protein. This study, however, was able to provide insight into the effects of serotonin binding on developing a preactive state of 5HT3A and how membrane lipids diversity influences protein structure and function.
Much simulation space to explore with these models remains, and it's only been made possible in the past few years with strides in computational power. Specifically, this study was performed using a grant-based allocation on the Anton2 supercomputer maintained by the Pittsburgh Supercomuting Center (PSC) and sponsored by DE Shaw research. Traditional high-performance computing resources would not be capable of reaching the timescales of this study.
As computing speed and power increases in the future, the group hopes to study the effects of pharmaceuticals, such as granisetron or ondansetron, in an effort to accurately model their effects on the protein and investigate other targeted therapies.

